Введение
Этот блог пост использует различные запросы к Большим Языковым Моделям (БЯМ) для суммаризации статьи “Искусство аттриционной войны: Уроки войны России против Украины” от Алекса Вершинина.
Замечание: Мы тоже будем пользоваться сокращением “LLM” (для “Large Language Models”).
В этой статье для Королевского института объединенных служб (RUSI), Алекс Вершинин обсуждает необходимость для Запада пересмотреть свою военную стратегию в отношении аттрициона в предвидении затяжных конфликтов. Статья противопоставляет аттриционную и маневренную войну, подчеркивая важность промышленной мощности, генерации сил и экономической устойчивости в победе в затяжных войнах.
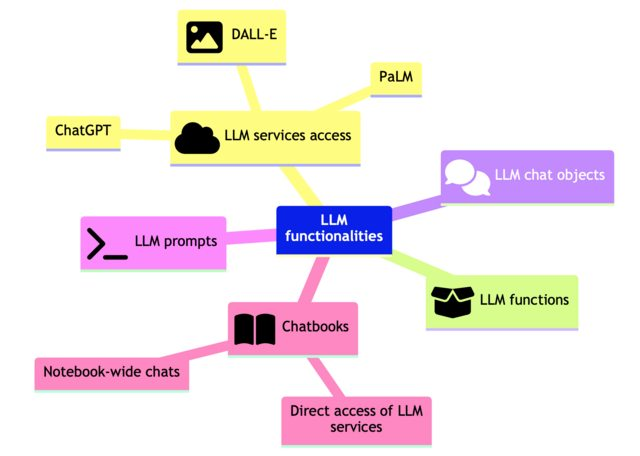
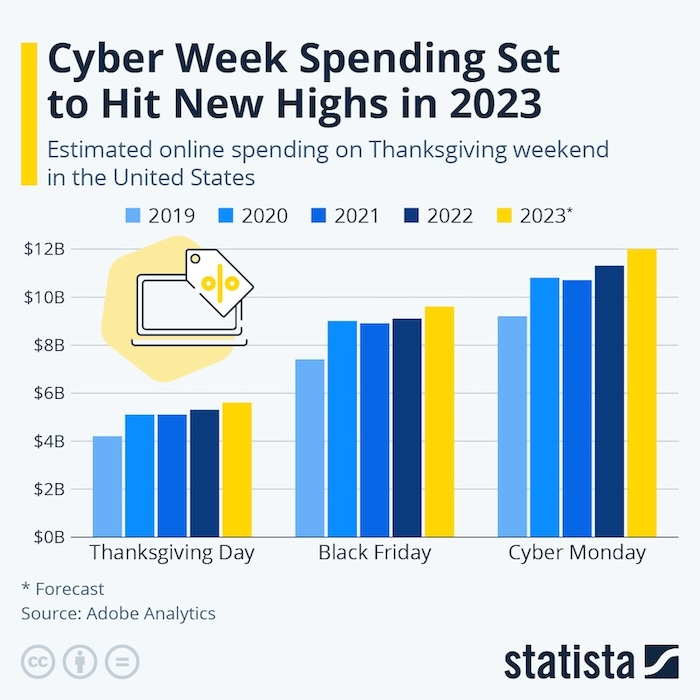
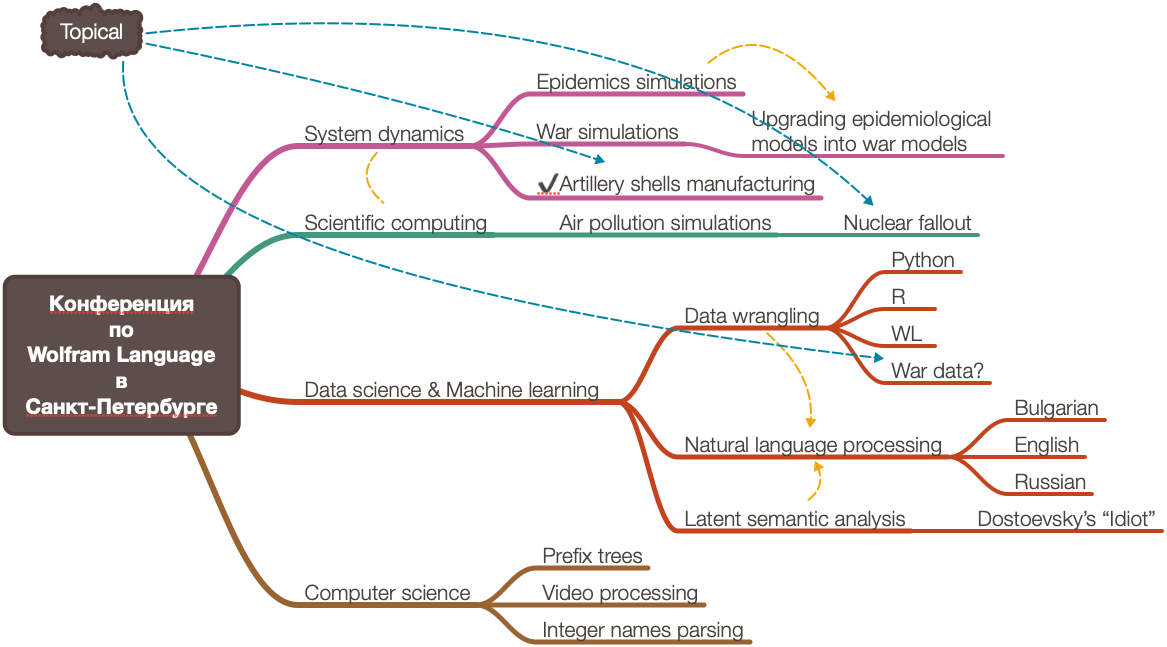
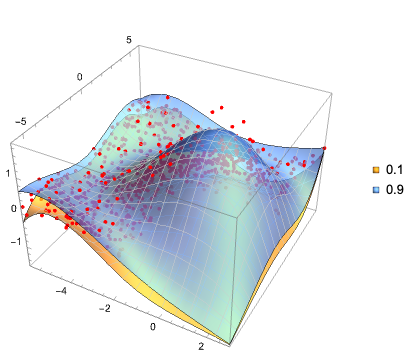
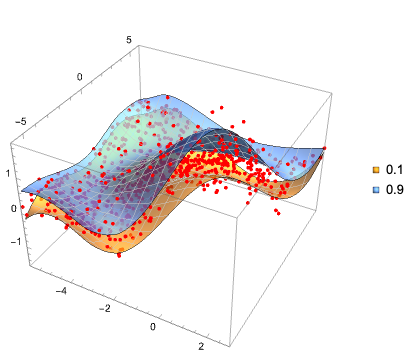
Эта (полученная с помощью LLM) иерархическая диаграмма хорошо суммирует статью:

Примечание: Мы планируем использовать этот пост/статью в качестве ссылки в предстоящем посте/статье с соответствующей математической моделью
(на основе Системной динамики.)
Структура поста:
- Темы
Табличное разбиение содержания. - Ментальная карта
Структура содержания и связи концепций. - Суммарное изложение, идеи и рекомендации
Основная помощь в понимании. - Модель системной динамики
Как сделать данный наблюдения операциональными?
Темы
Вместо суммарного изложения рассмотрите эту таблицу тем:
| тема | содержание |
|---|---|
| Введение | Статья начинается с подчеркивания необходимости для Запада подготовиться к аттриционной войне, контрастируя это с предпочтением коротких, решающих конфликтов. |
| Понимание Аттриционной Войны | Определяет аттриционную войну и подчеркивает ее отличия от маневренной войны, акцентируя важность промышленной мощности и способности заменять потери. |
| Экономическое Измерение | Обсуждает, как экономика и промышленные мощности играют ключевую роль в поддержании войны аттрициона, с примерами из Второй мировой войны. |
| Генерация Сил | Исследует, как различные военные доктрины и структуры, такие как НАТО и Советский Союз, влияют на способность генерировать и поддерживать силы в аттриционной войне. |
| Военное Измерение | Детализирует военные операции и стратегии, подходящие для аттриционной войны, включая важность ударов над маневрами и фазы таких конфликтов. |
| Современная Война | Исследует сложности современной войны, включая интеграцию различных систем и вызовы координации наступательных операций. |
| Последствия для Боевых Операций | Описывает, как аттриционная война влияет на глубинные удары и стратегическое поражение способности противника регенерировать боевую мощь. |
| Заключение | Резюмирует ключевые моменты о том, как вести и выигрывать аттриционную войну, подчеркивая важность стратегического терпения и тщательного планирования. |
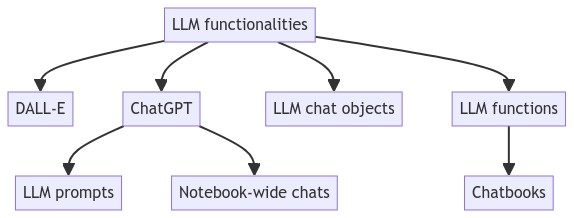
Ментальная карта
Вот ментальная карта показывает структуру статьи и суммирует связи между представленными концепциями:

Суммарное изложение, идеи и рекомендации
СУММАРНОЕ ИЗЛОЖЕНИЕ
Алекс Вершинин в “Искусстве аттриционной войны: Уроки войны России против Украины” для Королевского института объединенных служб обсуждает необходимость для Запада пересмотреть свою военную стратегию в отношении аттрициона в предвидении затяжных конфликтов.
Статья противопоставляет аттриционную и маневренную войну, подчеркивая важность промышленной мощности, генерации сил и экономической устойчивости в победе в затяжных войнах.
ИДЕИ:
- Аттриционные войны требуют уникальной стратегии, сосредоточенной на силе, а не на местности.
- Западная военная стратегия традиционно отдает предпочтение быстрым, решающим битвам, не готова к затяжному аттриционному конфликту.
- Войны аттрициона со временем выравнивают шансы между армиями с различными начальными возможностями.
- Победа в аттриционных войнах больше зависит от экономической силы и промышленной мощности, чем от военного мастерства.
- Интеграция гражданских товаров в военное производство облегчает быстрое вооружение в аттриционных войнах.
- Западные экономики сталкиваются с трудностями в быстром масштабировании военного производства из-за мирного эффективности и аутсорсинга.
- Аттриционная война требует массового и быстрого расширения армий, что требует изменения стратегий производства и обучения.
- Эффективность военной доктрины НАТО ухудшается в аттриционной войне из-за времени, необходимого для замены опытных некомиссированных офицеров (NCOs).
- Советская модель генерации сил, с ее массовыми резервами и офицерским управлением, более адаптируема к аттриционной войне.
- Соединение профессиональных сил с массово мобилизованными войсками создает сбалансированную стратегию для аттриционной войны.
- Современная война интегрирует сложные системы, требующие продвинутого планирования и координации, что затрудняет быстрые наступательные маневры.
- Аттриционные стратегии сосредоточены на истощении способности противника регенерировать боевую мощь, защищая свою собственную.
- Начальная фаза аттриционной войны подчеркивает удерживающие действия и наращивание боевой мощи, а не завоевание территории.
- Наступательные операции в аттриционной войне следует откладывать до тех пор, пока резервы и промышленная мощность противника достаточно не истощены.
- Глубинные удары по инфраструктуре и производственным возможностям противника имеют решающее значение в аттриционной войне.
- Аттриционная война требует стратегического терпения и акцента на оборонительных операциях для подготовки к будущим наступлениям.
- Ожидание Запада коротких, решающих конфликтов не соответствует реальности потенциальных аттриционных войн с равными противниками.
- Признание важности экономических стратегий и промышленной мобилизации ключево для подготовки к и выигрышу затяжного конфликта.
- Информационные операции могут манипулировать движениями и распределением ресурсов противника в свою выгоду в аттриционной войне.
ЦИТАТЫ:
- “Аттриционные войны требуют своего ‘Искусства войны’ и ведутся с ‘подходом, сосредоточенным на силе’.”
- “Та сторона, которая принимает аттриционный характер войны и сосредотачивается на уничтожении вражеских сил, а не на завоевании территории, скорее всего, победит.”
- “Войны аттрициона выигрываются экономиками, позволяющими массовую мобилизацию армий через их промышленные сектора.”
- “Проще и быстрее производить большое количество дешевого оружия и боеприпасов, особенно если их подкомпоненты взаимозаменяемы с гражданскими товарами.”
- “Эффективность военной доктрины НАТО ухудшается в аттриционной войне
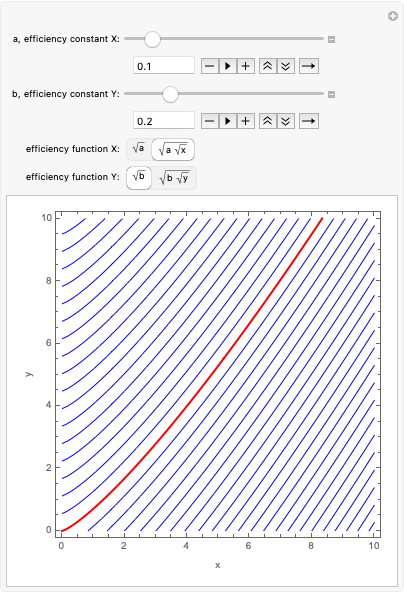
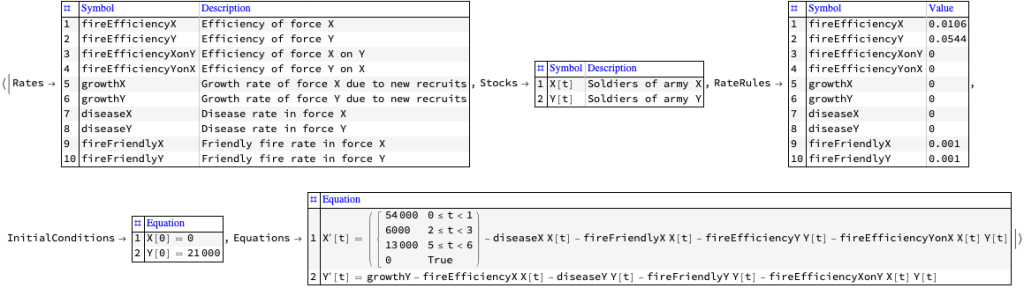
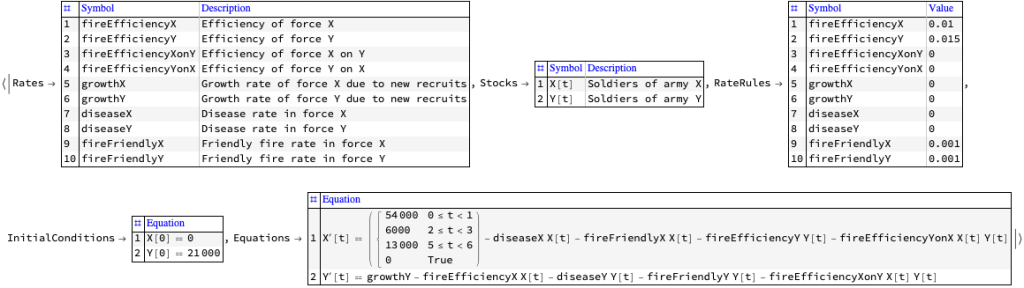
Модель системной динамики
В этом разделе мы показываем модель системной динамики, полученную с помощью запроса:
Сгенерируйте модель системной динамики для войны на истощение, описанной в следующей статье:
Описание модели, полученное с помощью LLM, имеет смысл, но для получения соответствующей рабочей модели требуются дополнительные запросы к LLM и/или (математическое) программирование и моделирование.
Результат LLM представлен ниже.
Разработка модели системной динамики для войны на истощение, описанной в предоставленном тексте, включает создание структуры, которая улавливает ключевые динамики истощения, генерации сил, экономической поддержки и влияния промышленной мощности на ведение войны. Эта модель будет сосредоточена на взаимодействиях между этими элементами для понимания долгосрочных результатов войны на истощение, особенно в контексте российской войны на Украине, как обсуждаетс
Компоненты модели системной динамики:
- Запасы: Это накопления в системе, которые изменяются со временем. Для модели войны на истощение актуальные запасы могут включать:
- Военный персонал: Сюда входят как высококвалифицированные профессиональные солдаты, так и мобилизованные или призванные военнослужащие низкого уровня.
- Промышленная мощность: Способность производить военное оборудование, которое может быть разделено на высокотехнологичные сложные системы и массово производимые предметы низкого уровня.
- Экономические ресурсы: Это финансовые и материальные ресурсы, доступные для поддержки военного производства и операций.
- Боевая мощь: Общая эффективность и сила военных сил, на которую влияют как количество, так и качество сил и оборудования.
- Потоки: Это скорости, которые изменяют запасы, представляя процессы внутри системы.
- Набор и обучение: Добавление личного состава к запасу военного персонала.
- Скорость истощения: Уменьшение военного персонала через боевые потери.
- Скорость производства: Скорость, с которой промышленная мощность производит военное оборудование.
- Распределение ресурсов: Поток экономических ресурсов в военное производство и операции.
- Петли обратной связи: Эти петли помогают понять, как разные части системы влияют друг на друга, усиливая или уравновешивая динамику системы.
- Усиливающая петля (R1): Увеличение промышленной мощности приводит к большему количеству военного оборудования, что повышает боевую мощь, потенциально приводя к военному успеху, который оправдывает дальнейшие инвестиции в промышленную мощность.
- Уравновешивающая петля (B1): Высокие скорости истощения сокращают военный персонал, снижая боевую мощь, что может привести к переоценке военных стратегий и потенциальному сокращению агрессивных операций для сохранения сил.
- Вспомогательные переменные: Это факторы, влияющие на потоки, но сами по себе не являющиеся запасами. Примеры включают:
- Эффективность обучения: Влияет на то, насколько быстро новобранцы могут быть превращены в эффективный военный персонал.
- Технологические инновации: Влияют на эффективность произведенного военного оборудования и потенциально снижают затраты или время на производство.
- Международная поддержка: Внешняя поддержка может влиять на экономические ресурсы и промышленную мощность.
- Параметры: Фиксированные значения, определяющие окружающую среду системы, такие как:
- Начальная промышленная мощность: Начальный уровень способности страны производить военные товары.
- Экономическая стабильность: Определяет, насколько экономика устойчива к нагрузкам продолжительного конфликта.
Процесс моделирования:
- Инициализация: Установка начальных значений для запасов на основе исторических данных или оценок (например, промышленная мощность до войны, начальный уровень военного персонала).
- Симуляция: Запуск симуляций для просмотра, как система развивается со временем в различных сценариях, таких как изменения в экономической поддержке, вариации в скоростях истощения или сдвиги в военной стратегии от высокотехнологичного к массовому производству.
- Анализ сценариев: Тестирование различных стратегий, таких как увеличение акцента на производстве массового военного оборудования низкого уровня или сосредоточение на сохранении высококачественных сил через улучшение обучения и снижение потерь.
- Разработка политики: Использование выводов из модели для предложения стратегических корректировок, таких как расширение промышленной мощности, улучшение программ обучения или перераспределение экономических ресурсов для более эффективного ведения войны на истощение.
Эта модель системной динамики предоставляет структурированный способ анализа сложных взаимодействий в войне на истощение, помогая стратегам и политикам понимать долгосрочные последствия различных военных и экономических стратегий.
Диаграмма модели системной динамики
Вот диаграмма описания модели, указанной выше:
Примечание: Приведенная выше диаграмма не представляет собой модель системной динамики как таковую. Она представляет концептуальные связи такой модели. В предстоящей статье мы планируем представить фактическую модель системной динамики с соответствующим описанием, диаграммами, уравнениями и результатами симуляции.