Introduction
In this notebook we show how to evaluate Machine Learning (ML) classifiers based on Tries with frequencies, [AA1, AA2, AAp1], created over a well known ML dataset. The computations are done with packages and functions from the Wolfram Language (WL) ecosystem.
The classifiers based on Tries with frequencies can be seen as generalized Naive Bayesian Classifiers (NBCs).
We use the workflow summarized in this flowchart:
Import["https://raw.githubusercontent.com/antononcube/MathematicaForPrediction/master/MarkdownDocuments/Diagrams/A-monad-for-classification-workflows/Classification-workflow-horizontal-layout.jpg"]
For more details on classification workflows see the article “A monad for classification workflows”, [AA3].
Remark: This notebook is the Mathematica counterpart of the Raku computable Markdown document with the same name [AA7, AA6].
Remark: Mathematica and WL are used as synonyms in this notebook.
Data
In this section we obtain a dataset to make classification experiments with.
Through the Wolfram Function Repository (WFR) function ExampleDataset we can get data for the Titanic ship wreck:
dsTitanic0 = ResourceFunction["ExampleDataset"][{"MachineLearning", "Titanic"}];
dsTitanic0[[1 ;; 4]]
Remark: ExampleDataset uses ExampleData. Datasets from the ExampleData’s “MachineLearning” and “Statistics” collections are processed in order to obtain Dataset objects.
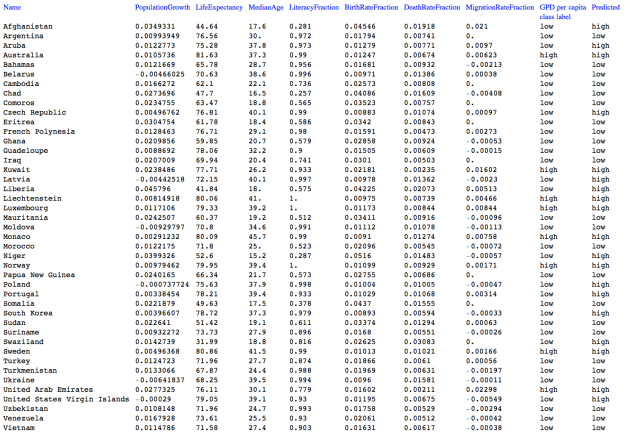
Instead of using the built-in Titanic data we use a version of it (which is used in Mathematica-vs-R comparisons, [AAr1]):
dsTitanic[[1 ;; 4]]
Here is a summary of the data:
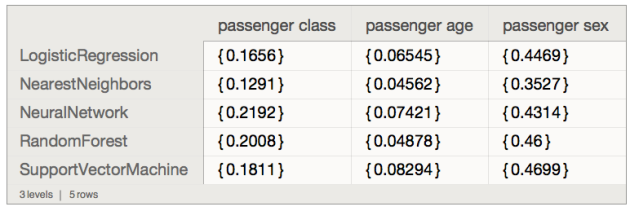
ResourceFunction["RecordsSummary"][dsTitanic]
Make tries
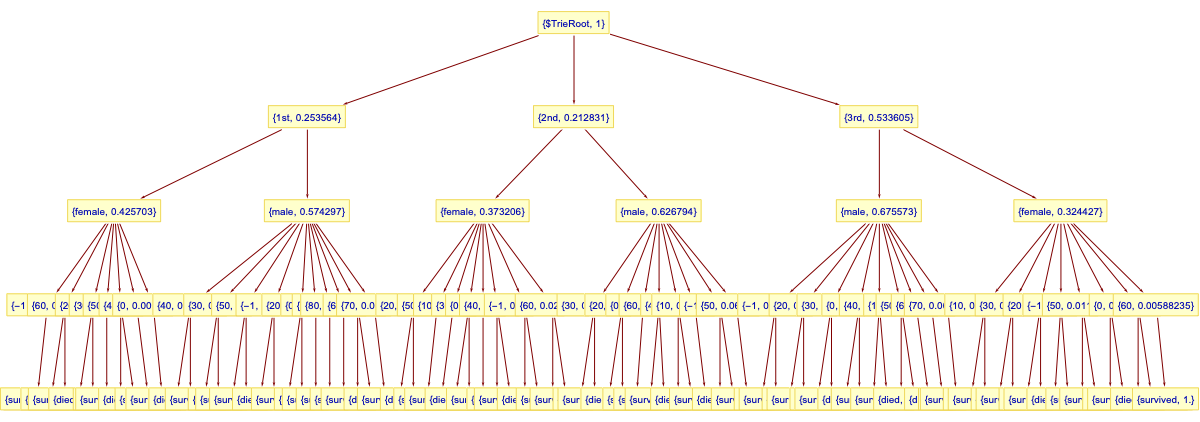
In this section for demonstration purposes let us create a shorter trie and display it in tree form.
Here we drop the identifier column and take the record fields in a particular order, in which “passengerSurvival” is the last field:
lsRecords = Normal@dsTitanic[All, Prepend[KeyDrop[#, "id"], "passengerAge" -> ToString[#passengerAge]] &][All, {"passengerClass", "passengerSex", "passengerAge", "passengerSurvival"}][Values];Here is a sample:
RandomSample[lsRecords, 3]
(*{{"3rd", "female", "-1", "died"}, {"1st", "female", "50", "survived"}, {"3rd", "male", "30", "died"}}*)Here make a trie without the field “passengerAge”:
trTitanic = TrieCreate[lsRecords[[All, {1, 2, 4}]]];
TrieForm[trTitanic, AspectRatio -> 1/4, ImageSize -> 900]
Here is a corresponding mosaic plot, [AA4, AAp3]:
MosaicPlot[lsRecords[[All, {1, 2, 4}]]]
Remark: The function MosaicPlot uses tries with frequencies in its implementation. Observing and reasoning with mosaic plots should make it clear that tries with frequencies are (some sort of) generalized Naive Bayesian classifiers.
Trie classifier
In this section we create a Trie-based classifier.
In order to make certain reproducibility statements for the kind of experiments shown here, we use random seeding (with SeedRandom) before any computations that use pseudo-random numbers. Meaning, one would expect WL code that starts with a SeedRandom statement (e.g. SeedRandom[89]) to produce the same pseudo random numbers if it is executed multiple times (without changing it.)
SeedRandom[12];Here we split the data into training and testing data in a stratified manner (a split for each label):
aSplit1 = GroupBy[lsRecords, #[[-1]] &, AssociationThread[{"training", "testing"}, TakeDrop[RandomSample[#], Round[0.75*Length[#]]]] &];
Map[Length, aSplit1, {2}]
(*<|"survived" -> <|"training" -> 375, "testing" -> 125|>, "died" -> <|"training" -> 607, "testing" -> 202|>|>*)Here we aggregate training and testing data (and show the corresponding sizes):
aSplit2 = <|
"training" -> Join @@ Map[#training &, aSplit1],
"testing" -> Join @@ Map[#testing &, aSplit1]|>;
Length /@ aSplit2
(*<|"training" -> 982, "testing" -> 327|>*)Here we make a trie with the training data (and show the node counts):
trTitanic = TrieNodeProbabilities[TrieCreate[aSplit2["training"]]];
TrieNodeCounts[trTitanic]
(*<|"total" -> 142, "internal" -> 60, "leaves" -> 82|>*)Here is the trie in tree form:
Here is an example decision-classification:
TrieClassify[trTitanic, {"1st", "female"}]
(*"survived"*)Here is an example probabilities-classification:
TrieClassify[trTitanic, {"1st", "female"}, "Probabilities"]
(*<|"survived" -> 0.962264, "died" -> 0.0377358|>*)We want to classify across all testing data, but not all testing data-records might be present in the trie. Let us check that such testing records are few (or none):
Tally[Map[TrieKeyExistsQ[trTitanic, #] &, aSplit2["testing"]]]
(*{{True, 321}, {False, 6}}*)Let us remove the records that cannot be classified:
lsTesting = Pick[aSplit2["testing"], Map[TrieKeyExistsQ[trTitanic, #] &, aSplit2["testing"]]];
Length[lsTesting]
(*321*)Here we classify all testing records and show a sample of the obtained actual-predicted pairs:
lsClassRes = {Last[#], TrieClassify[trTitanic, Most[#]]} & /@ lsTesting;
RandomSample[lsClassRes, 6]
(*{{"survived", "survived"}, {"died", "survived"}, {"died", "died"}, {"survived", "survived"}, {"survived", "died"}, {"survived", "survived"}}*)Here we cross tabulate the actual vs predicted labels using WFR’s CrossTabulate:
ResourceFunction["CrossTabulate"][lsClassRes]
The cross-tabulation results look bad because the default decision threshold is used. We get better results by selecting a decision threshold via Receiver Operating Characteristic (ROC) plots.
Trie classification with ROC plots
In this section we systematically evaluate the Trie-based classifier using the Receiver Operating Characteristic (ROC) framework.
Here we classify all testing data records. For each record:
- Get probabilities association
- Add to that association the actual label
- Make sure the association has both survival labels
lsClassRes = Map[Join[<|"survived" -> 0, "died" -> 0, "actual" -> #[[-1]]|>, TrieClassify[trTitanic, #[[1 ;; -2]], "Probabilities"]] &, lsTesting];Here we make a ROC record, [AA5, AAp4]:
ToROCAssociation[{"survived", "died"}, #actual & /@ lsClassRes, Map[If[#survived >= 0.5, "survived", "died"] &, lsClassRes]]
(*<|"TruePositive" -> 71, "FalsePositive" -> 14, "TrueNegative" -> 184, "FalseNegative" -> 52|>*)Here we obtain the range of the label “survived”:
lsVals = Map[#survived &, lsClassRes];
MinMax[lsVals]
(*{0, 1.}*)Here we make list of decision thresholds:

In the following code cell for each threshold:
- For each classification association decide on “survived” if the corresponding value is greater or equal to the threshold
- Make threshold’s ROC-association
lsROCs = Table[
ToROCAssociation[{"survived", "died"}, #actual & /@ lsClassRes, Map[If[#survived >= th, "survived", "died"] &, lsClassRes]],
{th, lsThresholds}];Here is the obtained ROCs dataset:
Dataset[MapThread[Prepend[#1, "Threshold" -> #2] &, {lsROCs, lsThresholds}]]
Here is the corresponding ROC plot:
ROCPlot["FPR", "TPR", lsThresholds, lsROCs, GridLines -> Automatic, ImageSize -> Large]
We can see the Trie-based classifier has reasonable prediction abilities – we get ≈ 80% True Positive Rate (TPR) for a relatively small False Positive Rate (FPR), ≈ 20%.
Confusion matrices
Using ClassifierMeasurements we can produce the corresponding confusion matrix plots (using “made on the spot” Manipulate interface):
DynamicModule[{lsThresholds = lsThresholds, lsClassRes = lsClassRes, lsClassRes2},
Manipulate[
lsClassRes2 = Map[{#actual, If[#survived >= lsThresholds[[i]], "survived", "died"]} &, lsClassRes];
Append[DeleteCases[ClassifierMeasurements[lsClassRes2[[All, 1]], lsClassRes2[[All, 2]], "ConfusionMatrixPlot"], ImageSize -> _], ImageSize -> Medium],
{{i, Flatten[Position[lsThresholds, Nearest[lsThresholds, 0.3][[1]]]][[1]], "index:"}, 1, Length[lsThresholds], 1, Appearance -> "Open"},
{{normalizeQ, False, "normalize?:"}, {False, True}}
]
]
References
Articles
[AA1] Anton Antonov, “Tries with frequencies for data mining”, (2013), MathematicaForPrediction at WordPress.
[AA2] Anton Antonov, “Tries with frequencies in Java”, (2017), MathematicaForPrediction at WordPress.
[AA3] Anton Antonov, “A monad for classification workflows”, (2018), MathematicaForPrediction at WordPress.
[AA4] Anton Antonov, “Mosaic plots for data visualization”, (2014), MathematicaForPrediction at WordPress.
[AA5] Anton Antonov, “Basic example of using ROC with Linear regression”, (2016), MathematicaForPrediction at WordPress.
[AA6] Anton Antonov, “Trie based classifiers evaluation”, (2022), RakuForPrediction-book at GitHub/antononcube.
[AA7] Anton Antonov, “Trie based classifiers evaluation”, (2022), RakuForPrediction at WordPress.
Packages
[AAp1] Anton Antonov, TriesWithFrequencies Mathematica package, (2014-2022), MathematicaForPrediction at GitHub/antononcube.
[AAp2] Anton Antonov, ROCFunctions Mathematica package, (2016), MathematicaForPrediction at GitHub/antononcube.
[AAp3] Anton Antonov, MosaicPlot Mathematica package, (2014), MathematicaForPrediction at GitHub/antononcube.
Repositories
[AAr1] Anton Antonov, Mathematica vs. R project, (2018-2022), GitHub/antononcube.
Setup
Import["https://raw.githubusercontent.com/antononcube/MathematicaForPrediction/master/TriesWithFrequencies.m"];
Import["https://raw.githubusercontent.com/antononcube/MathematicaForPrediction/master/ROCFunctions.m"];
Import["https://raw.githubusercontent.com/antononcube/MathematicaForPrediction/master/MosaicPlot.m"];dsTitanic = Import["https://raw.githubusercontent.com/antononcube/MathematicaVsR/master/Data/MathematicaVsR-Data-Titanic.csv", "Dataset", HeaderLines -> 1];